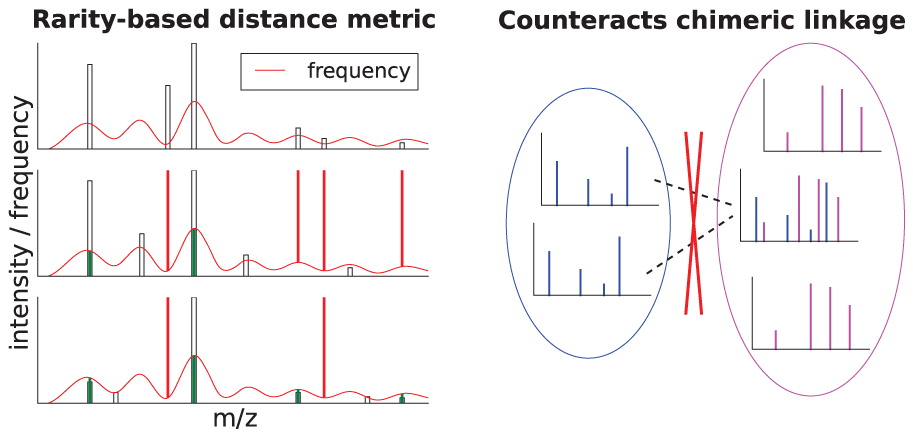
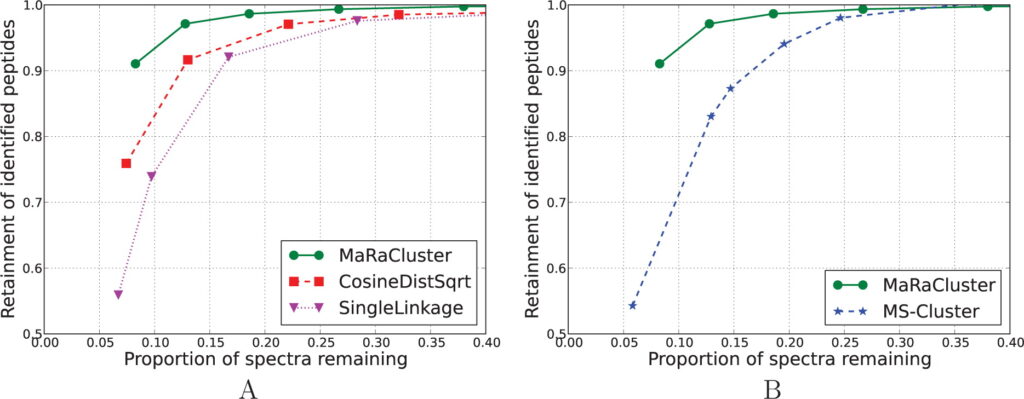
Large-scale proteomics datasets contain MS2 spectra that appear repeatedly across experiments, creating thousands of redundant spectra. This wastes computation and obscures weaker signals. MaRaCluster (Mass Rarity Cluster) solves this by grouping similar spectra based on how rare or common their fragment peaks are. Peaks that appear infrequently carry more weight, making the clustering highly selective. Once clustered, it creates consensus spectra that are cleaner and easier for search engines to identify. This not only speeds up database searches but also boosts identification rates, especially for low-abundance peptides. By turning spectral redundancy into a strength, MaRaCluster helps researchers dig deeper into their data with less effort.
Examples, binaries and installation guides are available on the GitHub page.
Github: https://github.com/statisticalbiotechnology/maracluster
Publication: The, M., Käll, L. (2016). MaRaCluster: A Fragment Rarity Metric for Clustering Fragment Spectra in Shotgun Proteomics. J. Proteome Res.,