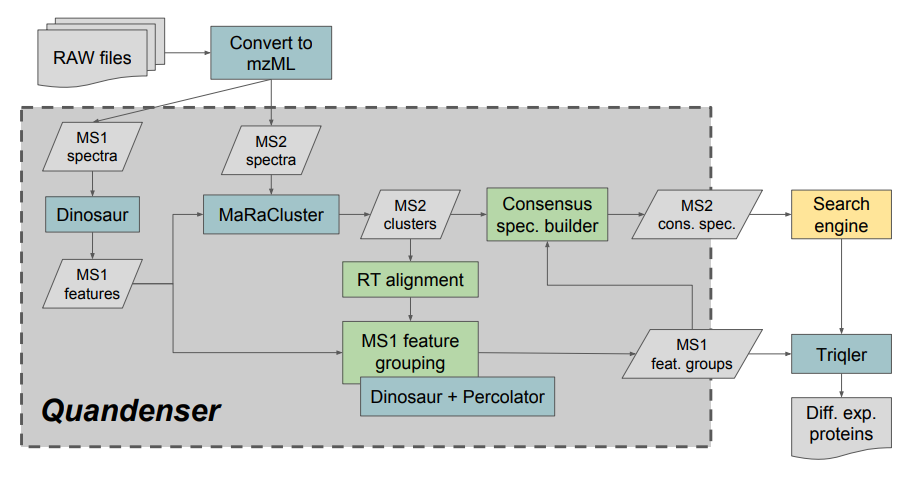
Mass-spec proteomics is drowning in data: many spectra go unused, and traditional pipelines discard a lot of raw quantitative signal before identification. Quandenser (QUANtification by Distillation for ENhanced Signals with Error Regulation) reverses that logic. It first clusters both MS1 (precursor) features and MS2 (fragment) spectra in an unsupervised way, condensing the data into “feature groups” before attempting identification. This quantification-first strategy uncovers analytes that would otherwise be ignored, reduces computational load, and enables open-modification or de novo searches on condensed spectra. Crucially, Quandenser estimates error rates for feature matching (e.g. match-between-runs) and feeds them into Triqler’s Bayesian quantification framework. When paired (Quandenser + Triqler), the method finds more true differences between conditions without compromising error control.
Examples, binaries and installation guides are available on the GitHub page.
Github: https://github.com/statisticalbiotechnology/quandenser
Publication: The, M., Käll, L. (2020). Focus on the spectra that matter by clustering of quantification data in shotgun proteomics. Nature Communications,