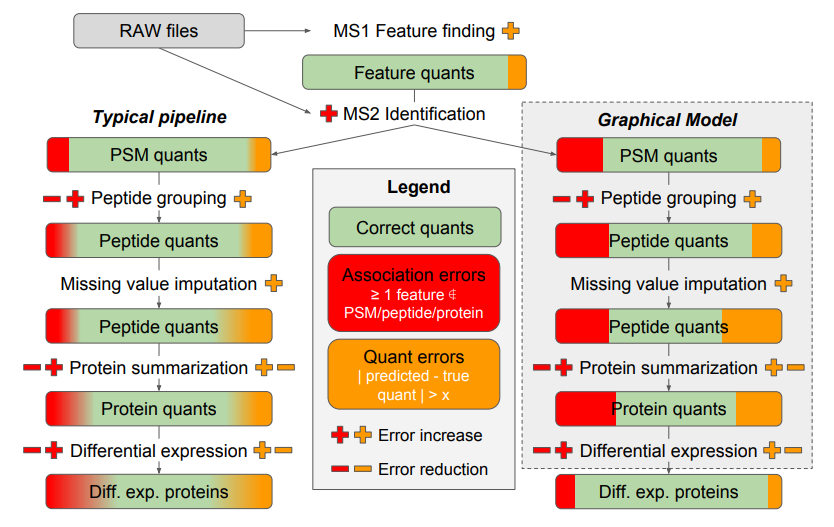
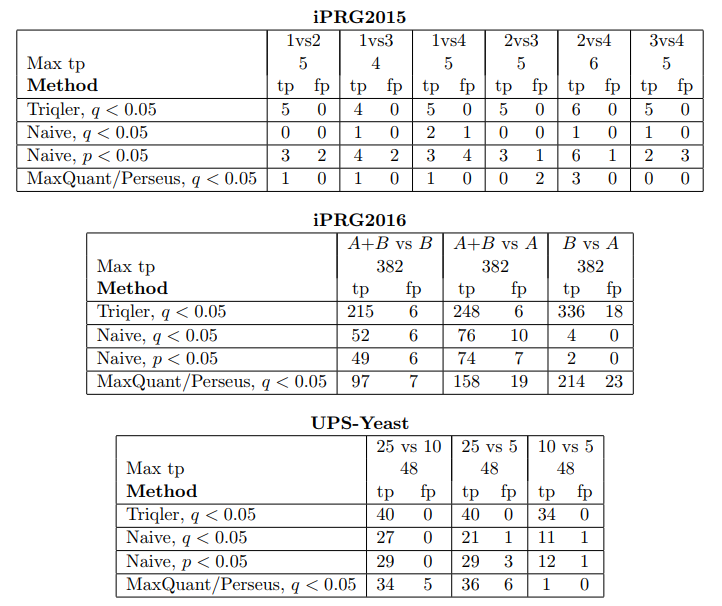
Proteomics is full of uncertainty: missing values, noisy peptide intensities and ambiguous identifications. Most tools sweep these issues under the rug by applying hard thresholds or imputation. Triqler (TRansparent Identification-Quantification-linked Error Rates) does the opposite. It uses a probabilistic model that links identification and quantification errors into a single Bayesian framework. Instead of forcing binary decisions, it calculates the probability that a protein is truly different between conditions. This approach reduces false positives without sacrificing sensitivity. For biologists, it means more trustworthy discoveries. For bioinformaticians, it means principled error propagation instead of ad-hoc filters. In short, Triqler turns messy data into meaningful conclusions.
Triqler is a Python package and can be installed easily using pip. Examples and installation guides are available on the GitHub page.
Github: https://github.com/statisticalbiotechnology/triqler
Publication: The, M., Käll, L. (2019). Integrated identification and quantification error probabilities for shotgun proteomics. Molecular & Cellular Proteomics,